The Programme funds and provides support to several different projects into childhood cancer research.
Barbieri group
The Barbieri group, based in the Department of Pathology at the University of Cambridge, are investigating the role of RNA modification in paediatric leukaemias and lymphomas, particularly through METTL1, METTL3 and TGS1.
Latest publications
Miano et al. The non-coding epitranscriptome in cancer. Brief Funct Genomics. March 2021
Barbieri & Kouzarides. Role of RNA modifications in cancer. Nat Rev Cancer. April 2020
Pandolfini et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell April 2019
Pump Prime projects 2020-2021
The Paediatric Cancer Programme is currently funding 2 pump prime projects:
| Use of cellular barcoding to track the origins of metastatic tumour cells in Ewing’s Sarcoma | Prof Matthew Allen, Dept of Veterinary Medicine |
| NPM-ALK is an oncogenic hyperactive kinase that directs transcription via modification of chromatin | Dr Suzanne Turner, Dept of Pathology |
Use of cellular barcoding to track the origins of metastatic tumour cells in Ewing’s Sarcoma
Prof Matthew Allen, Department of Veterinary Medicine | £15,000 | March 2020 – April 2021
Collaborators: Dr Isaia Barbieri, Dept of Pathology
Background to this project
In recent work with a mouse model of Ewing’s sarcoma, the Allen group at the Department of Veterinary Medicine, has validated a new approach for tracking and isolating tumour cells from the primary site in bone, the peripheral circulation and metastatic sites in the lung.
This new project will see the Allen group collaborate with Dr Isaia Barbieri and his group at the Department of Pathology to incorporate a cellular barcoding approach.
Project goals
The goal of this project is to combine a metastasis tracking approach with a cellular barcoding strategy that will allow for accurate identification of parent cells in the primary Ewing’s sarcoma tumour that are the source of metastasis.
This will allow researchers to:
- Determine the engraftment capacity of these cells in vivo
- Better understand the clonal selection process that occurs upon metastasis formation
- Determine whether consistent patterns in clonal selection are occurring, if so, assisting with the design of single-cell RNA-seq studies to identify key molecular drivers or suppressors of metastasis
What does this mean for patients?
Longer term, this project aims to kick start the development of a validated panel of gene markers for risk of disease progression, providing physicians with a much better ability to accurately identify patients at high risk of metastasis and to focus treatment and post treatment monitoring.
This gene panel will also help researchers to identify new approaches in the design of less toxic therapies. Depending on the nature of the gene product identified as a driver or suppressor of metastasis, there may be an option to selectively target the expression of bioavailability of the gene product in patients.
Background to Ewing’s sarcoma
Ewing’s sarcoma is an aggressive form of bone cancer that is seen predominantly in teenagers and young adults. Even with aggressive treatment, most often including surgery and chemotherapy, many patients will succumb to the disease when it metastasises to other organs such as the lung. The overall 5-year survival rate for patients with Ewing’s sarcoma is around 50%, a figure that has not changed appreciably in the last two decades.
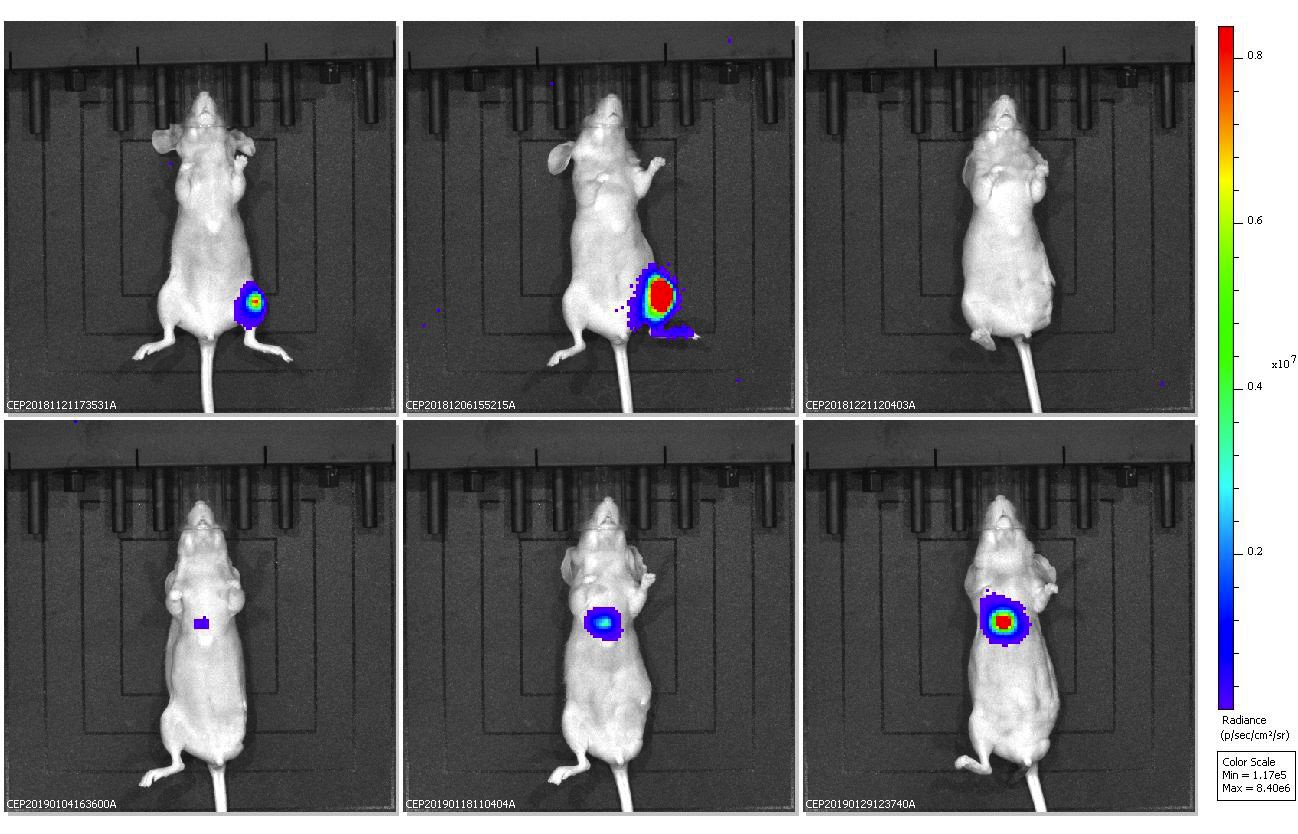
Figure: Bioluminescence imaging demonstrates growth and metastasis of TC71 Ewing’s sarcoma cells in nude mice. After initial primary growth in the tibia (weeks 2-4), the signal is lost at 6 weeks following amputation, but then returns as lung metastases develop (weeks 8-12).
NPM-ALK is an oncogenic hyperactive kinase that directs transcription via modification of chromatin
Dr Suzanne Turner, Department of Pathology | £15,000 | March 2020 – August 2021
Collaborators: Dr Isaia Barbieri, Dept of Pathology
Background to this project
NPM-ALK, generated as the consequence of a chromosomal translocation, is the driving oncogenic event in Anaplastic Large Cell Lymphoma (ALCL). Most studies of NPM-ALK activity to date have focused on events in the cytoplasm despite its presence also in the nucleus. The nuclear activity of this kinase is therefore poorly understood.
In this project the Turner group will collaborate with Dr Isaia Barbieri and his group to investigate the role of NPM-ALK in chromatin modification and activation of transcription. The project will also involve collaborations with Namshik Han at the Milner Institute for bioinformatic data analyses, and with Megan Lim at the University of Pennsylvania for mass spectrometry.
Project goals
- To investigate the role of NPM-ALK as a histone kinase
- To establish the role of NPM-ALK as a director of transcription
What does this mean for patients?
Longer term, further understanding of the role of NPM-ALK in ALCL and the role of epigenetics in disease will help researchers target new treatments towards processes that are fundamental to disease.
Background to Anaplastic Large Cell Lymphoma (ALCL)
Anaplastic Large Cell Lymphoma is a cancer affecting mostly children and resulting from the transformation of thymocytes in the developing thymus.
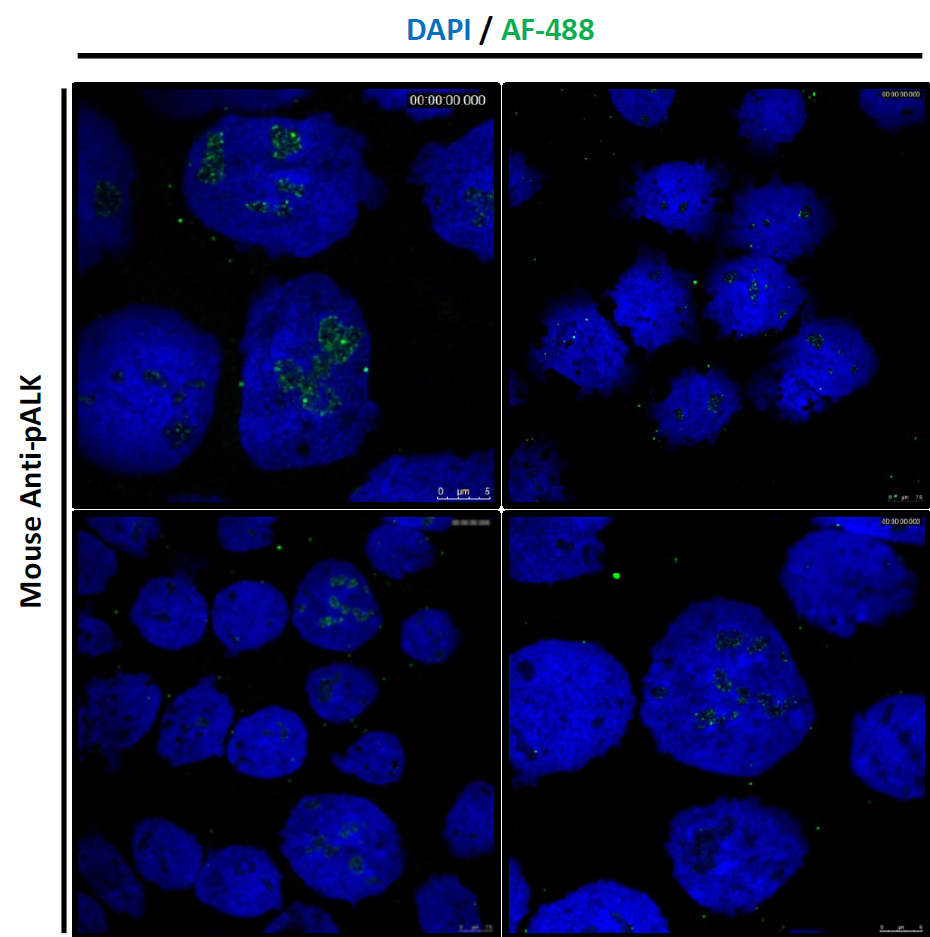
Figure: Confocal microscopy shows the nucleolar localisation of active, tyrosine phosphorylated NPM-ALK in cancer cells