Services offered by CMDL

Whole blood processing for isolation of:
- Plasma
- Serum
- Buffy coat
- PBMCs
- Granulocytes
Processing of other body fluids, including:
- Saliva
- Ascites
- Urine
- Bone marrow aspirate
- Pleural fluid

Manual and automated (QIAsymphony, Maxwell RSC 48, Biomek FXP) nucleic acid extractions, genomic DNA (gDNA), circulating cell free DNA (ccfDNA) and RNA, from:
- Plasma
- Serum
- Bone marrow aspirate
- Buffy coat
- Granulocytes
- Fresh frozen tissue
- PBMCs
- Ascites
- Pleural fluid
- Urine
- Saliva
- FFPE tissue slides/scrolls
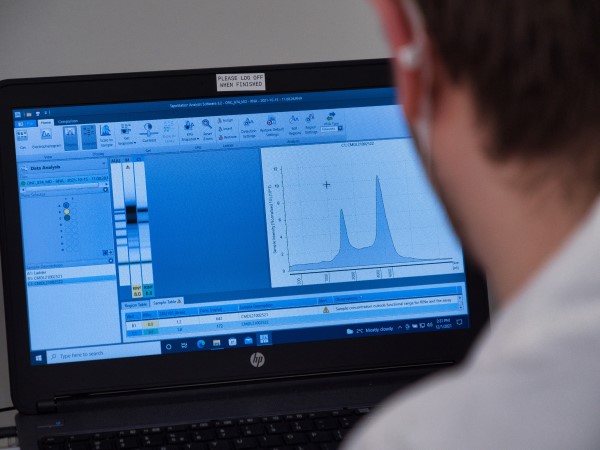
- Nucleic acid quantification: Qubit, Fluoroskan, qPCR
- Nucleic acid fragment size analysis: 2100 Bioanalyzer, 4200 TapeStation
- Cellular quantification and viability: LUNA FL

We offer a spectrum of validated NGS approaches.
We are always open to trialling and adopting new approaches to accommodate a study.
Currently available panels validated for gDNA, FFPE gDNA and ccfDNA:
- 350 gene pan-cancer panel (Twist Bioscience Custom Capture Panel)
- 110 gene lymphoid malignancy panel (Twist Bioscience Custom Capture Panel)
- 56 gene myeloid panel for myeloid malignancies and clonal haematopoesis (Twist Bioscience Custom Capture Panel)
- Shallow whole genome sequencing (sWGS)
- T cell receptor (TCR) sequencing (QIAseq Immune Repertoire RNA Library Kit)
- Ampliseq Cancer Hotspot Panel (commercial Illumina amplicon based panel for actionable variants in hotspot regions of oncogenes)
Library preparation is performed at CMDL and sequencing is performed by SMCL.
For all our NGS workflows, we offer an in-house bioinformatics analysis which includes quality control and variant calling as standard, following Genome Analysis Toolkit's Best Practices.
Standard fastq files are generated from the internal bcl Illumina files using Illumina’s own bcl2fastq package with all the recommended settings. Data is then returned via a password protected https link.
We currently offer assays for:
- Real-time PCR approaches including methylation analysis of the MGMT gene promoter
- QX200 Droplet Digital PCR system for variant detection and copy number analysis assays, among many
- NanoString nCounter assays on request (mainly gene expression and CNV analysis from RNA)
This video explains the steps required to process whole blood samples into buffy coat and double-spun plasma samples. This process is routinely followed in the CMDL to generate blood product samples for cancer research.
This video is intended as a visual training aide alongside full procedure documentation, risk assessment, and in-person training. No liability is accepted when following this training video alone. Please always follow the appropriate safety precautions when handling sharps, blood, and blood products.



















