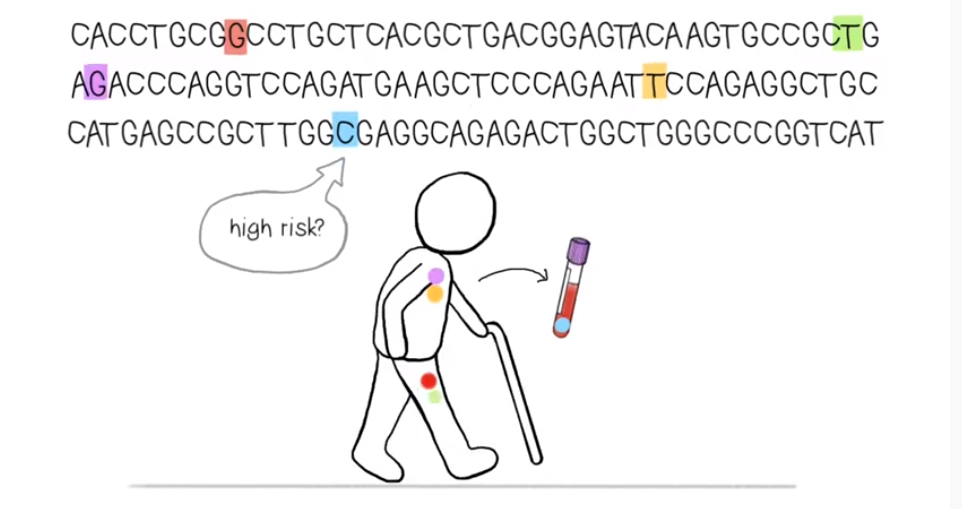
In the animation above, lead author Dr Caroline Watson explains the results of research, published in the journal Science, studying patterns of mutations in healthy blood samples to predict the risk of developing cancer.
Cancer is evolution at the cellular level.
As we age, we acquire mutations in the cells that make up our tissues.
Whilst the vast majority of these mutations are harmless, some of them increase the Darwinian ‘fitness’ of cells, allowing them to expand and outcompete our healthy cells.
These expansions increase the chance of accumulating further cancer-causing mutations and can thus increase the risk of developing cancer.
A major question in the field of early cancer detection is therefore: which specific mutations enable cells to expand most rapidly, and thus might confer the highest risk of cancer?
Blood’s relative ease of sampling has resulted in the generation of a huge amount of genomic data in recent years and this provides researchers with an ideal opportunity to answer this question.
Led by Dr Jamie Blundell, researchers in our Early Detection Programme have developed a method that is able to estimate the growth rates of mutations that expand in healthy tissues, without needing to use longitudinal data.
Dr Blundell said: “Blood provides an ideal model system for understanding the earliest stages in cancer development. The scale and resolution of the genomic data in blood is unparalleled, and, combined with quantitative methods borrowed from evolutionary biology has enabled us to identify the variants most likely to pose a high risk of a future blood cancer developing.”
The research team applied evolutionary theory to mutation size estimates in blood sequencing data collected from around 50,000 individuals to quantify the growth potential (or ‘fitness effect’) of specific mutations at single nucleotide resolution.
This enabled them to build a league table of the ‘fittest’ and therefore potentially most pathogenic mutations in blood.
The researchers were also able to quantify the distribution of fitness effects within genes and therefore what proportion of mutations within a gene are potentially high risk.
Lead author Dr Caroline Watson commented: "Knowing whether specific mutations are high-risk or clinically insignificant will be key in the future of personalised cancer risk stratification. Our framework provides a rational basis for quantifying the growth potential of mutations and, in combination with studies that can track these mutations and their outcomes over time, will be an important step towards this goal.”
Combining their framework with future studies that longitudinally track individuals over time will shed light on how mutations drive the development of cancer and will help to accelerate the development of risk predictors.
Reference
Caroline J. Watson, A. L. Papula, Gladys Y. P. Poon, Wing H. Wong, Andrew L. Young, Todd E. Druley, Daniel S. Fisher, Jamie R. Blundell
The evolutionary dynamics and fitness landscape of clonal hematopoiesis Science, 27 March 2020.















