About us
DNA-damage response science in radiotherapy
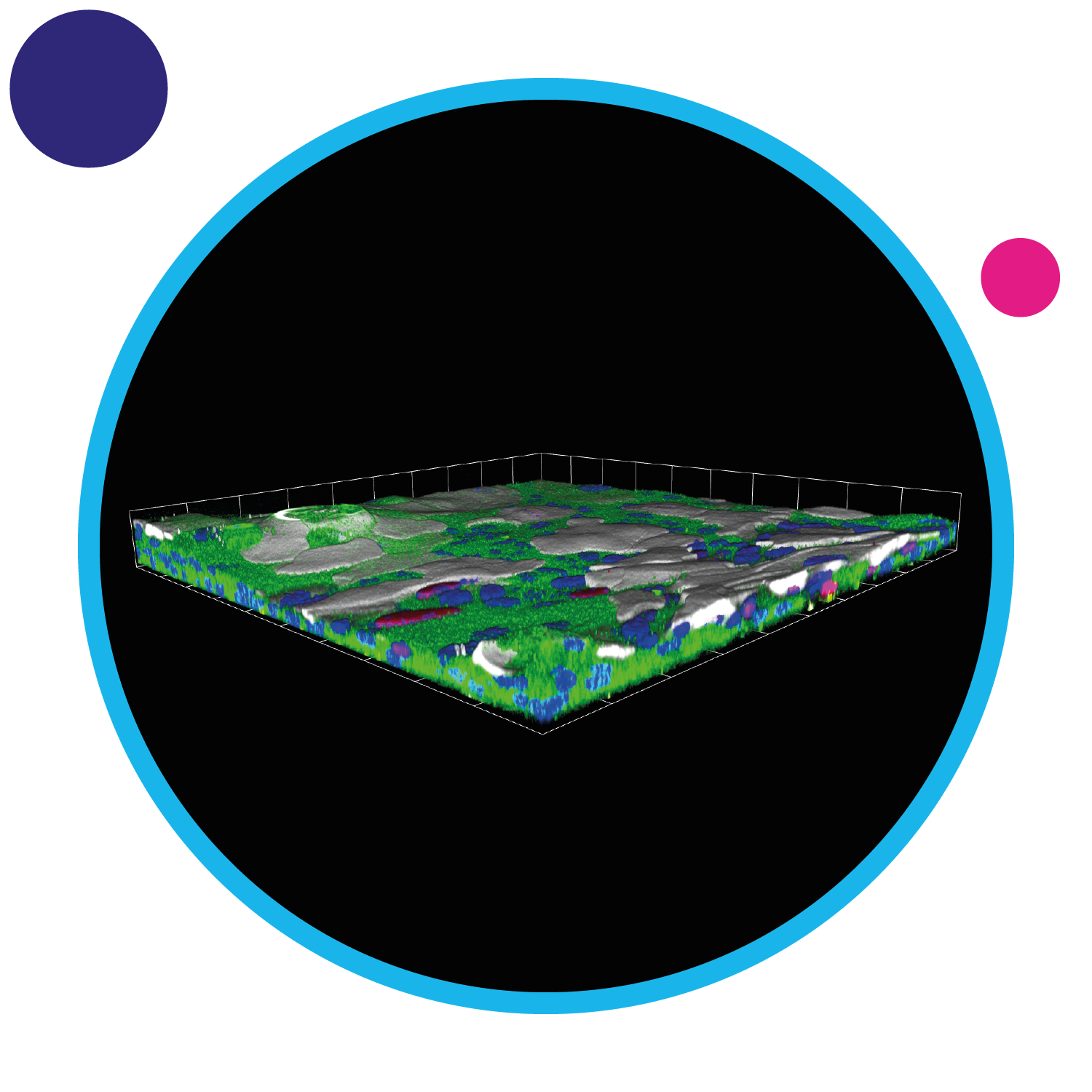
We are applying our innovative 3D culture system for mouse and human primary tissues (a system known as epithelioid culture) to understand the molecular mechanisms by which normal cells and mutant cancer progenitor cells show different responses to radiotherapy treatments. We are studying the role of some mutations, such as NOTCH1 and TP53, in oral and oesophageal normal epithelia during the very early events of tumour formation.
We are also involved in the characterisation of the FLASH effect in normal and tumour tissues together with Professor Karen Kirby (Manchester) and Dr Kristoffer Petersson (Oxford). FLASH treatment is based on delivering radiotherapy at ultra-high dose-rates (typically greater than 40 Gy/s) and it appears to offer increased normal tissue sparing, in comparison to conventional dose rate radiotherapy, while still suppressing tumour growth. This work will investigate a broad range of physical (dose, dose-rate, radiation quality, and pulse structure) and hypoxic conditions which will be critical in determining the optimum parameters to realise FLASH in the clinic. Now we are analysing the differential induction of DNA double strand breaks between conventional, FLASH proton, and FLASH electron radiotherapies.

Novel technology to screen for radiotherapy-drug targets
This theme utilises Cambridge's world-leading expertise in functional genomic approaches, where we systematically disrupt the genome to discover effectors of radiosensitivity and resistance in the treatment of cancer. We investigate these genetic susceptibilities not just in isolation, but also in the presence of DNA-damage response inhibitors. By using clinically-relevant, radiation-resistant models of cancer in vitro and in vivo, we are trying to find new drug targets to improve patient outcomes when treating some of the most radiation-resistant cancers.
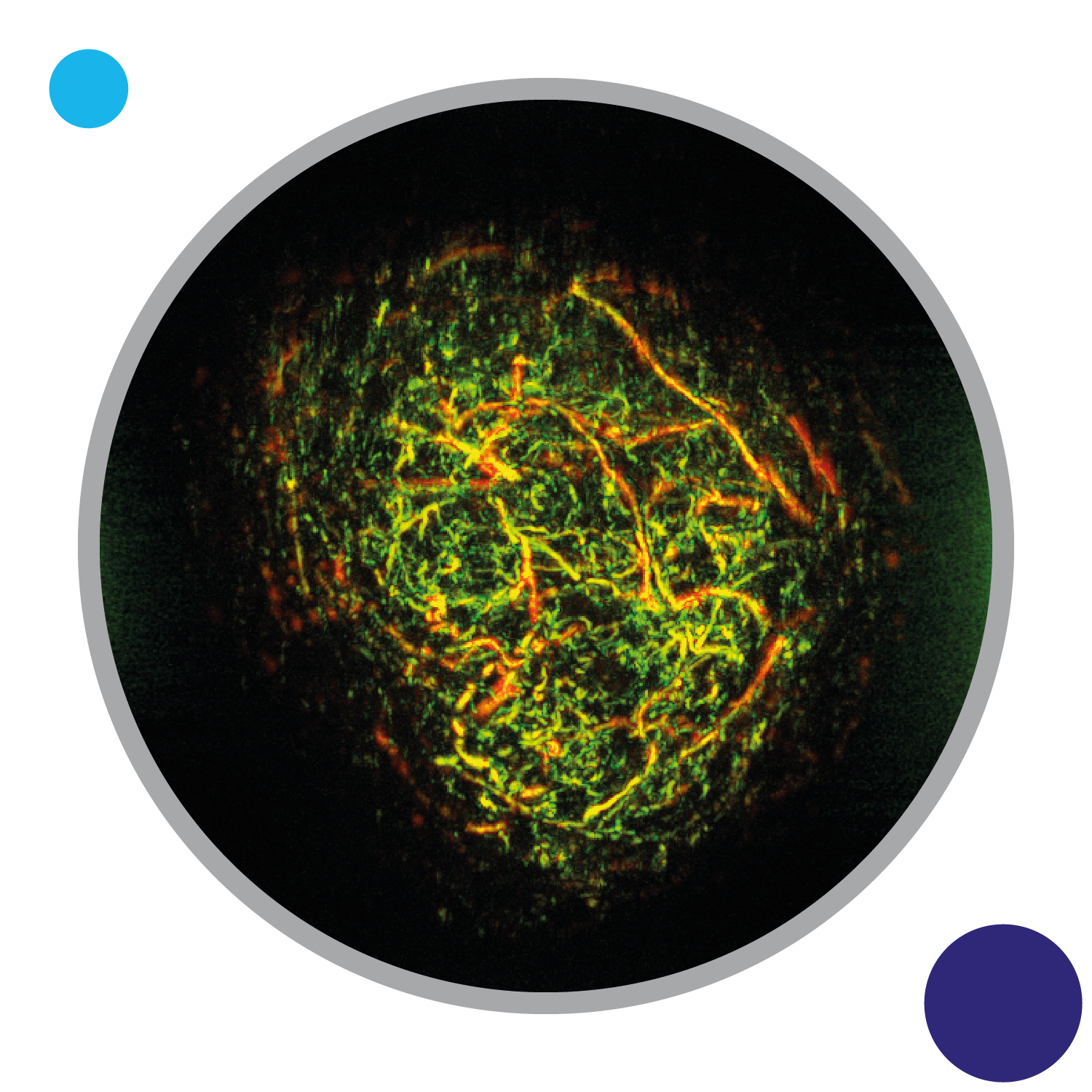
Evaluating clinically-relevant radiotherapy tumour models
This theme is focused on creating cancer models and methods that help us to robustly test new radiotherapy strategies. We are testing different ways of combining radiotherapy with chemotherapy for improved outcomes. We apply state-of-the-art imaging methods to watch tumours responding to treatment, and are also creating new computational analyses to understand how we can enhance response to radiotherapy and mitigate resistance.

Designing translationally-rich radiotherapy clinical trials
The focus of this theme is to learn as much as possible from every patient recruited into our portfolio of advanced radiotherapy studies, by integrating bio-sample collection into studies for translational research. For example, Hamlet.rt trans is a sub-study where patients undergoing radiotherapy for lung or head and neck cancer give weekly samples of their blood. We then use these samples in cutting-edge digital PCR techniques to look for signals of tumour radiation response during the course of radiotherapy.
We are combining this analysis of circulating tumour DNA with concurrent analysis of blood-based protein factors that correlate with the senescence response of tumour and healthy tissues. Some tumour cells can be pushed into a 'sleeping' state (called senescence) after radiotherapy. Regrowth of these tumour cells at a later time can lead to recurrence of the tumour. Newly-developed drug therapies known as senolytics can be given to target these sleeping tumour cells after radiotherapy treatment.
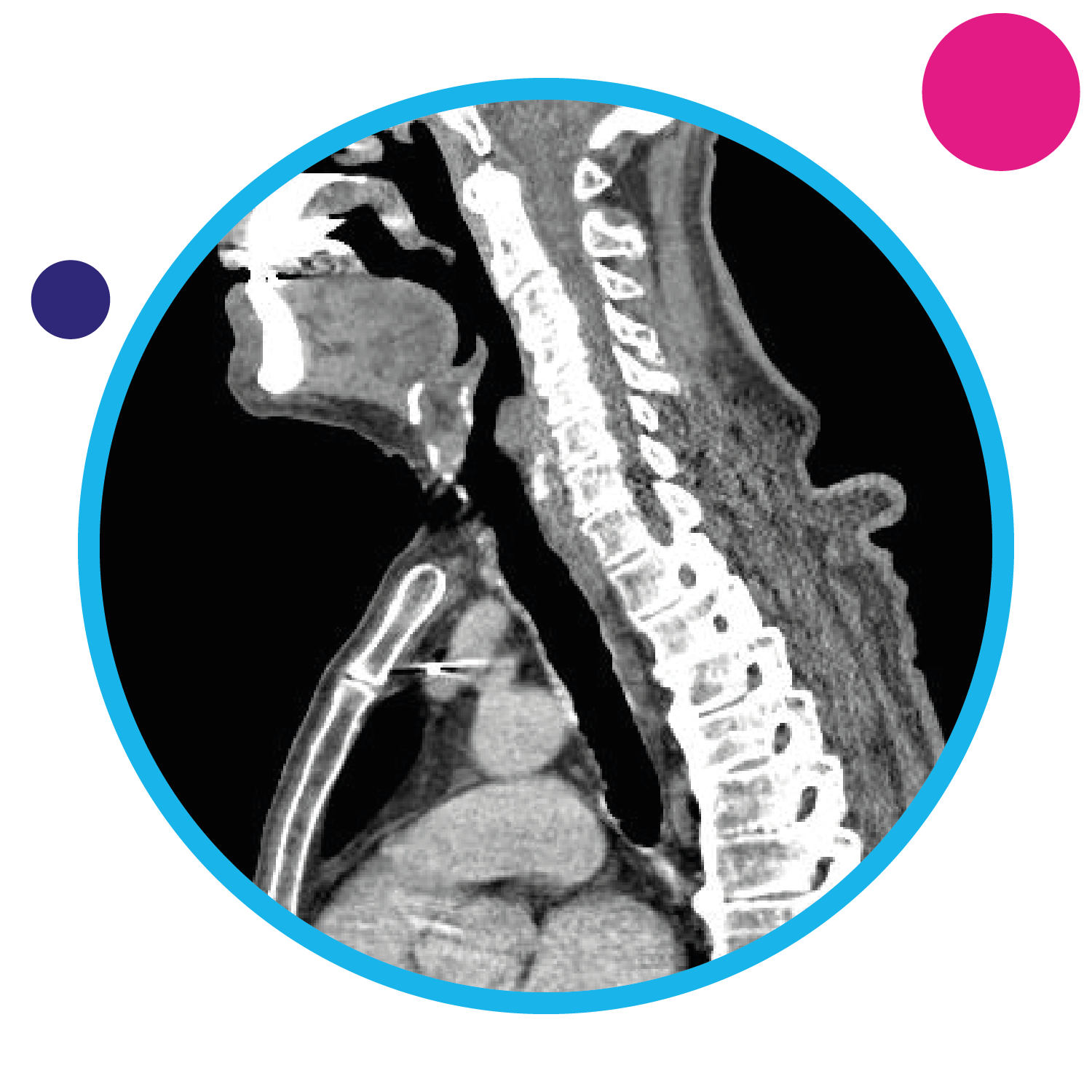
Machine learning, and predictive radiomics and genomics for radiotherapy
This theme uses artificial intelligence to obtain new insights from patients who have been recruited into practice-defining radiotherapy trials. We use the power of data science and machine learning to analyse imaging data and patient outcome data, alongside their genomic data, to understand the factors that determine the control of tumours, patterns of disease recurrence, and the side effect profile of radiotherapy. Our work has been used to create Hamlet.rt, a multi-centre prospective study for machine learning in radiotherapy and a platform for translational research.
















