Dr Michael Boemo
Position: Lecturer
Personal home page:
https://www.boemogroup.org/
Email:
mb915@cam.ac.uk
PubMed journal articles - click here
Dr Michael Boemo is pleased to consider applications from prospective PhD students.
My group develops new computational methods to study how dysregulation and errors in DNA replication and mitosis lead to genome instability. We use and develop cutting-edge methodologies from machine learning, bioinformatics, software engineering, high-performance computing, and computational modelling to realise this goal.
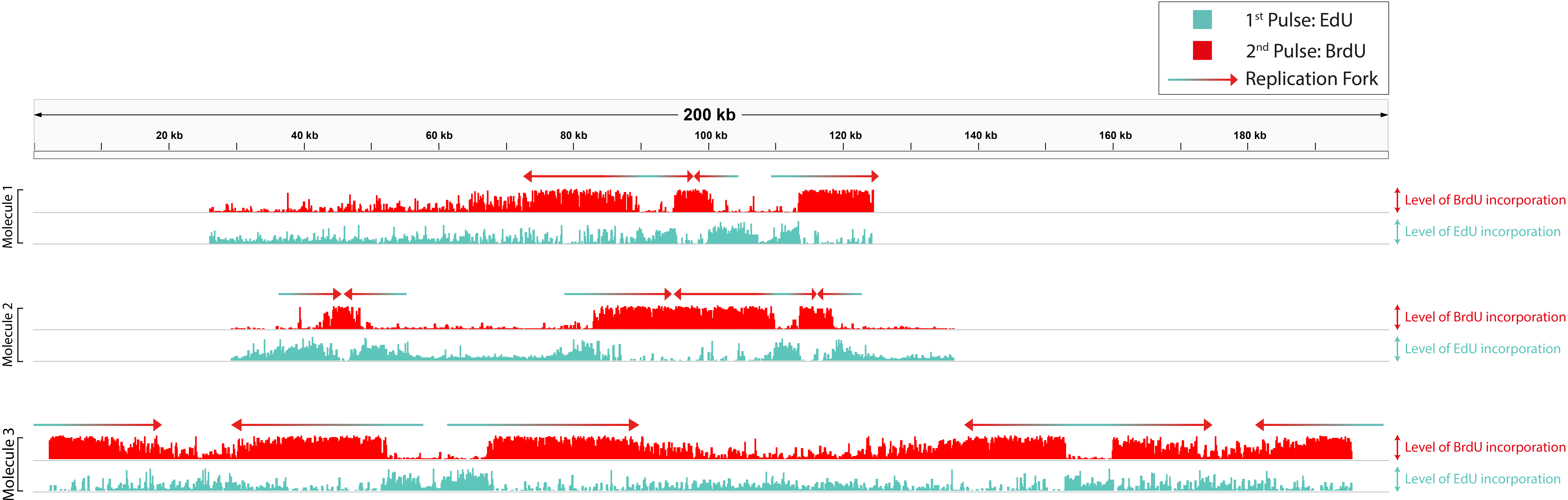
Before each human cell divides, six billion base pairs of DNA have to be replicated quickly and accurately. This enormous task is accomplished by the asynchronous, distributed action of tens of thousands of replication forks. Errors in this process can occur when replication forks stall on encountering an obstacle that they cannot pass, such as DNA lesions or actively transcribed genes. When left unresolved, stalled forks can leave a region of the genome unreplicated, potentially resulting in the copy number alterations, structural rearrangements, and micronuclei that underpin genome instability. While frequent fork stalling is thought to drive oncogenesis, DNA replication pathways are an attractive therapeutic target whereby fork stalling is induced by a number of chemotherapeutic agents currently used in the clinic. Where, why, and how often replication forks stall in the genome is an unsolved question with important consequences for understanding oncogenesis and the mechanism of action of currently used treatments.
We are addressing this issue by developing machine learning software that infers replication initiation and fork movement by detecting base analogues in ultra-long Oxford Nanopore reads. When these analogues are pulsed into replicating DNA, they are incorporated into newly copied DNA by replication forks leaving a "footprint" of fork movement. The software is able to determine how and where replication forks were moving during the pulse. We partner with a range of experimental collaborators to address central questions pertaining to the regulation, performance, and behaviour DNA replication to understand how it affects genome integrity.
Computational Biology
Machine Learning
Bioinformatics
Systems Biology
Mathematical Modelling
DNA Replication
DNA Repair
Genome Stability
Symplectic Elements feed provided by Research Information, University of Cambridge
Jones, M.J.K.†, Rai, S.K., Pfuderer, P.L., Bonfim-Melo, A., Pagan, J.K., Clarke, P.R., McClelland, S.E., Boemo, M.A.† (2022) A high-resolution, nanopore-based artificial intelligence assay for DNA replication stress in human cancer cells. [preprint available on bioRxiv]
Shaikh, N.*, Mazzagatti, A.*, Bakker, B., Spierings, D.C.J.E., Wardenaar, R., Maniati, E., Larsson, P., Wang, J., Boemo, M.A., Foijer, F., McClelland, S.E.† (2022) DNA replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses. Genome Biology 23:223.
Boemo, M.A.† (2021) DNAscent v2: Detecting replication forks in nanopore sequencing data with deep learning. BMC Genomics 22:430.
Aydogan, M.G.*†, Steinacker, T.L.*, Mofatteh, M., Wilmott, Z.M., Zhou, F.Y., Gartenmann, L., Wainman, A., Saurya, S., Novak, Z.A., Wong, S., Goriely, A., Boemo, M.A.†, Raff, J.W.† (2020) A free-running oscillator times and executes centriole biogenesis. Cell 181:1-16.
Boemo, M.A.†, Cardelli, L., Nieduszynski, C.A. (2020) The Beacon Calculus: A formal method for the flexible and concise modelling of biological systems. PLoS Computational Biology 16:e1007651.
Mueller, C.A.*, Boemo, M.A.*, Spingardi, P., Kessler, B. Kriaucionis, S. Simpson, J.T., Nieduszynski, C.A.† (2019) Capturing the dynamics of genome replication on individual ultra-long nanopore sequencing reads. Nature Methods 16:429-436.
Boemo, M.A.†, Byrne, H.M.† (2018) Mathematical modelling of a hypoxia-regulated oncolytic virus delivered by tumour-associated macrophages. Journal of Theoretical Biology 461:102-116.